Papers Page Code
From GersteinInfo
(→[ Return to index]) |
|||
| (126 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | == | + | ==Word Cloud== |
| - | + | ||
| - | [ | + | Link to [[Word Cloud]] |
| - | == | + | == (Lab Member) Papers Page Rebuild and Further Info (Private)== |
| + | *Private Wiki includes instructions about how to update papers page and further info, click here [http://wiki.gersteinlab.org/labinfo/Papers_Page_Documentation Private Wiki] | ||
| - | ==== | + | ==Papers GitHub== |
| + | Papers 2.0 is now version controlled under GitHub: https://github.com/gersteinlab/papers.gersteinlab.org | ||
| + | ==SpreadSheet Structure== | ||
| + | *"Papers Page" is built up from two basic google spreadsheets, "Papers Master" and "Papers Subjects". "Papers Master" contains descriptions about papers, such as, pubmed ID, authors, citation, et al.. Papers are affiliated to some grants. Grants information is stored in "Papers Subjects". | ||
| + | Here is a list of the tags and their meanings<br \> | ||
| + | ===Papers Master=== | ||
| + | Papers Master: | ||
| + | <labid> - id by which to refer to the article | ||
| + | <PMID> - PubMed id | ||
| + | <title> - title of the article | ||
| + | <citation> - citation of the article (author, journal, year, etc) | ||
| + | <preprint> - URL of the preprint file | ||
| + | <subjects> - specifies the grant(s) funding the paper (e.g. "cegs,keck") | ||
| + | <website> - supplemental website | ||
| + | <Year> - year the article was published | ||
| + | <footnote> - additional information | ||
| + | <website2> - second supplemental website | ||
| - | + | The tags can conceptually be divided into two groups: ones such as PMID and title, which serve to identify the paper, and tags such as website and subject which supply supplemental information about the paper. There are two ways to identify a paper (in order of decreasing precedence): | |
| - | + | I. PMID<br \> | |
| + | II. title, citation | ||
| - | + | You should always include the PMID if a paper is known to be listed in PubMed. Option 2 should be used for papers that are in press. | |
| - | < | + | The other group of tags supplies additional information about the paper specified by the first group of tags. All of these tags are optional, however used of <subjects> and <preprint> is strongly encouraged. |
| - | + | {| border="1" cellpadding="0" cellspacing ="0" | |
| - | + | ! scope="col" width="50" align="center" style="background:#f0f0f0;"|'''labid''' | |
| - | + | ! scope="col" width="50" align="center" style="background:#f0f0f0;"|'''PMID''' | |
| - | + | ! scope="col" width="50" align="center" style="background:#f0f0f0;"|'''title''' | |
| - | + | ! scope="col" width="50" align="center" style="background:#f0f0f0;"|'''citation''' | |
| - | + | ! scope="col" width="50" align="center" style="background:#f0f0f0;"|'''preprint''' | |
| - | + | ! scope="col" width="50" align="center" style="background:#f0f0f0;"|'''subject''' | |
| - | + | ! scope="col" width="50" align="center" style="background:#f0f0f0;"|'''website''' | |
| - | + | ! scope="col" width="50" align="center" style="background:#f0f0f0;"|'''Year''' | |
| - | + | ! scope="col" width="50" align="center" style="background:#f0f0f0;"|'''footnote''' | |
| - | + | ! scope="col" width="100" align="center" style="background:#f0f0f0;"|'''website2''' | |
| - | + | |- | |
| - | + | |metamembrane||20430783||||||http://archive.gersteinlab.org/papers/e-print/metamembrane/preprint.pdf||interactions||http://metagenomics.gersteinlab.org/membrane||2010|||||| | |
| - | + | |} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | + | Spreadsheet of Paper IDs and PubMed IDs with other annotation: MBGLab--Papers-Master ([https://docs.google.com/spreadsheet/pub?hl=en_US&hl=en_US&key=0AiiHlTECOi8edGhzSXlZZzEyRThQeDB6R0pRc0FvcGc&output=html HTML]) ([https://docs.google.com/spreadsheet/pub?hl=en_US&hl=en_US&key=0AiiHlTECOi8edGhzSXlZZzEyRThQeDB6R0pRc0FvcGc&output=csv CSV]) | |
| - | + | ||
| - | + | ||
| - | + | ===Papers Subjects=== | |
| + | Papers Subjects: | ||
| + | <category> - classification of grants | ||
| + | <labid> - id refer to each grant | ||
| + | <title> - description of each grant | ||
| + | <website> - external website | ||
| + | <html> - additional information | ||
| - | + | We encourage you to sort <category> after adding new grants because of coding issues. <website> should also be reflected in <html> section. For example, "don" has <website> "http://www.donaghue.org", also "URL: <A HREF=http://www.donaghue.org> http://www.donaghue.org</A>" in <html> section. | |
| - | ''' | + | {| border="1" cellpadding="0" cellspacing ="0" |
| + | ! scope="col" width="50" align="center" style="background:#f0f0f0;"|'''category''' | ||
| + | ! scope="col" width="50" align="center" style="background:#f0f0f0;"|'''labid''' | ||
| + | ! scope="col" width="50" align="center" style="background:#f0f0f0;"|'''title''' | ||
| + | ! scope="col" width="50" align="center" style="background:#f0f0f0;"|'''website''' | ||
| + | ! scope="col" width="50" align="center" style="background:#f0f0f0;"|'''html''' | ||
| + | |- | ||
| + | |Research Grants||don||Dongahue Young Investigator||http://www.donaghue.org/||Young investigator award from Donaghue Foundation to M Gerstein (PI), "Comparative Genomics of Microbial Pathogens," (DF98-113, 1/1/99-12/31/03). URL: <A HREF=http://www.donaghue.org> http://www.donaghue.org</A>Articles funded by this grant: | ||
| + | |} | ||
| - | + | [https://docs.google.com/spreadsheet/pub?key=0AiiHlTECOi8edDBCUUhtMFZzRFVMaVlxZ1JrNGp1b0E&output=html HTML table of all subject headings in the current papers page] | |
| - | + | ||
| - | + | ===MBGLab--Papers XML Import=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [https://docs.google.com/spreadsheet/pub?key=0AiiHlTECOi8edHFxQWRwN0kxeTQ3ZzdCOXhQX2Z1ZGc&output=html HTML table of all PubMed entries in tabular format] | |
| - | + | ==Generate publication documents from SpreadSheet== | |
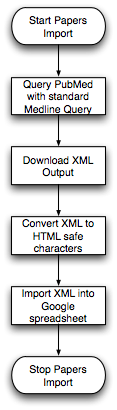
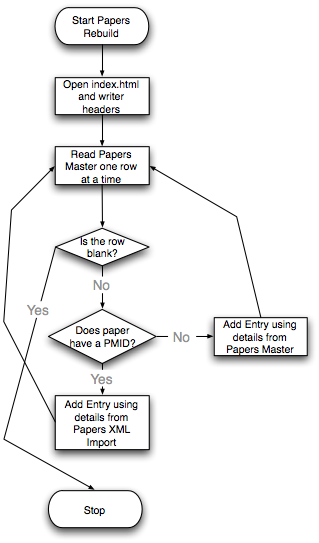
| - | + | Two Steps Flowchart (created by Mike Wilson): | |
| - | + | ||
| - | == | + | [[Image:Papers1.png||Import XML Flow]] [[Image:Papers2.png||Rebuild Papers Page]] |
| - | + | ||
| - | + | * Download XML file from NCBI using PubMed ID to generate pubmed_spreadsheet. Pubmed_spreadsheet stores <title> <citation> et al. of papers corresponding to "PMID" in "Papers Master". This step is done by scripts automatically. | |
| - | + | GoogleSpreadsheet.py: grab googlespreadsheet with python, see [[Grab_GoogleSpreadsheet_with_a_Python]]<br \> | |
| - | + | Other Code: see [[PubmedSpreadsheet_Generation_Code]] | |
| + | |||
| + | Pipeline: | ||
| + | First obtain pubmed_result.xml from papers medline query | ||
| + | parse_pmids.py<br \> curl `cat ncbiquery.txt` > NCBIData.xml | ||
| + | Reformat NCBIData.xml to tab delimited file to upload to Google | ||
| + | python import.py | ||
| + | replace PubMed Import XML spreadsheet with export_out.tab | ||
| + | reload_data.py | ||
| + | |||
| + | |||
| + | *Build Papers Page | ||
| + | This step grabs all information from three spreadsheets, "Papers Master","Papers Subjects" and "Pubmed_Spreadsheet", to build up the whole website. Each paper and each grant has its own description page. | ||
| + | |||
| + | Script used in this step: | ||
| + | |||
| + | update.py : see [[Build_Papers_Page_Code]] | ||
| + | |||
| + | ==Papers Page Code (Old)== | ||
| + | *[[Papers_Page_Code_Old]] | ||
| + | |||
| + | ==Old Papers Server ... and also R code for NSF colab == | ||
| + | *Old version of pagers website [http://oldpapers.gersteinlab.org Old Papers] | ||
| + | |||
| + | *Other info of papers [[Paper_search]] | ||
| + | |||
| + | R Code for compiling list of NSF co-authors: | ||
| + | This is buggy and approximate, but a good place to start | ||
| + | |||
| + | https://github.com/ejfertig/NSFBiosketch/blob/master/CollaboratorList.Rmd | ||
| - | == | + | library('easyPubMed') |
| - | + | library('plyr') | |
| + | library('xlsx') | ||
| + | author <- 'Gerstein M' | ||
| + | authorFilter <- 'Gerstein Maya' | ||
| + | currentDate <- Sys.Date() | ||
| + | queryDate <- seq(Sys.Date(), length = 2, by = "-48 months")[2] | ||
| + | pmquery <- sapply(articles_to_list(fetch_pubmed_data(get_pubmed_ids(paste0(author,'[Author]')))),article_to_df) | ||
| + | names(pmquery) <- NULL | ||
| + | pmquery <- pmquery[sapply(pmquery,function(x){any(colnames(x)=='year')})] | ||
| + | datePMID <- as.Date(sapply(pmquery,function(x){paste(x[1,'year'], x[1,'month'], x[1,'day'],sep="-")})) | ||
| + | pmquery <- pmquery[datePMID >= queryDate] | ||
| + | pmquery.dataframe <- ldply(pmquery,data.frame) | ||
| + | pmquery.dataframe$Initials <- getInitials(pmquery.dataframe$firstname) | ||
| + | pmquery.dataframe$ColabType <- 'Co-Author' | ||
| + | pmquery.dataframe <- pmquery.dataframe[!duplicated(paste(pmquery.dataframe$lastname,pmquery.dataframe$Initials)),] | ||
| + | pmquery.dataframe <- pmquery.dataframe[order(pmquery.dataframe$lastname,pmquery.dataframe$Initials),] | ||
Latest revision as of 17:02, 16 September 2019
Contents |
Word Cloud
Link to Word Cloud
(Lab Member) Papers Page Rebuild and Further Info (Private)
- Private Wiki includes instructions about how to update papers page and further info, click here Private Wiki
Papers GitHub
Papers 2.0 is now version controlled under GitHub: https://github.com/gersteinlab/papers.gersteinlab.org
SpreadSheet Structure
- "Papers Page" is built up from two basic google spreadsheets, "Papers Master" and "Papers Subjects". "Papers Master" contains descriptions about papers, such as, pubmed ID, authors, citation, et al.. Papers are affiliated to some grants. Grants information is stored in "Papers Subjects".
Here is a list of the tags and their meanings
Papers Master
Papers Master:
<labid> - id by which to refer to the article <PMID> - PubMed id <title> - title of the article <citation> - citation of the article (author, journal, year, etc) <preprint> - URL of the preprint file <subjects> - specifies the grant(s) funding the paper (e.g. "cegs,keck") <website> - supplemental website <Year> - year the article was published <footnote> - additional information <website2> - second supplemental website
The tags can conceptually be divided into two groups: ones such as PMID and title, which serve to identify the paper, and tags such as website and subject which supply supplemental information about the paper. There are two ways to identify a paper (in order of decreasing precedence):
I. PMID
II. title, citation
You should always include the PMID if a paper is known to be listed in PubMed. Option 2 should be used for papers that are in press.
The other group of tags supplies additional information about the paper specified by the first group of tags. All of these tags are optional, however used of <subjects> and <preprint> is strongly encouraged.
| labid | PMID | title | citation | preprint | subject | website | Year | footnote | website2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| metamembrane | 20430783 | http://archive.gersteinlab.org/papers/e-print/metamembrane/preprint.pdf | interactions | http://metagenomics.gersteinlab.org/membrane | 2010 |
Spreadsheet of Paper IDs and PubMed IDs with other annotation: MBGLab--Papers-Master (HTML) (CSV)
Papers Subjects
Papers Subjects:
<category> - classification of grants <labid> - id refer to each grant <title> - description of each grant <website> - external website <html> - additional information
We encourage you to sort <category> after adding new grants because of coding issues. <website> should also be reflected in <html> section. For example, "don" has <website> "http://www.donaghue.org", also "URL: <A HREF=http://www.donaghue.org> http://www.donaghue.org</A>" in <html> section.
| category | labid | title | website | html |
|---|---|---|---|---|
| Research Grants | don | Dongahue Young Investigator | http://www.donaghue.org/ | Young investigator award from Donaghue Foundation to M Gerstein (PI), "Comparative Genomics of Microbial Pathogens," (DF98-113, 1/1/99-12/31/03). URL: <A HREF=http://www.donaghue.org> http://www.donaghue.org</A>Articles funded by this grant: |
HTML table of all subject headings in the current papers page
MBGLab--Papers XML Import
HTML table of all PubMed entries in tabular format
Generate publication documents from SpreadSheet
Two Steps Flowchart (created by Mike Wilson):
- Download XML file from NCBI using PubMed ID to generate pubmed_spreadsheet. Pubmed_spreadsheet stores <title> <citation> et al. of papers corresponding to "PMID" in "Papers Master". This step is done by scripts automatically.
GoogleSpreadsheet.py: grab googlespreadsheet with python, see Grab_GoogleSpreadsheet_with_a_Python
Other Code: see PubmedSpreadsheet_Generation_Code
Pipeline:
First obtain pubmed_result.xml from papers medline query parse_pmids.py
curl `cat ncbiquery.txt` > NCBIData.xml Reformat NCBIData.xml to tab delimited file to upload to Google python import.py replace PubMed Import XML spreadsheet with export_out.tab reload_data.py
- Build Papers Page
This step grabs all information from three spreadsheets, "Papers Master","Papers Subjects" and "Pubmed_Spreadsheet", to build up the whole website. Each paper and each grant has its own description page.
Script used in this step:
update.py : see Build_Papers_Page_Code
Papers Page Code (Old)
Old Papers Server ... and also R code for NSF colab
- Old version of pagers website Old Papers
- Other info of papers Paper_search
R Code for compiling list of NSF co-authors: This is buggy and approximate, but a good place to start
https://github.com/ejfertig/NSFBiosketch/blob/master/CollaboratorList.Rmd
library('easyPubMed')
library('plyr')
library('xlsx')
author <- 'Gerstein M'
authorFilter <- 'Gerstein Maya'
currentDate <- Sys.Date()
queryDate <- seq(Sys.Date(), length = 2, by = "-48 months")[2]
pmquery <- sapply(articles_to_list(fetch_pubmed_data(get_pubmed_ids(paste0(author,'[Author]')))),article_to_df)
names(pmquery) <- NULL
pmquery <- pmquery[sapply(pmquery,function(x){any(colnames(x)=='year')})]
datePMID <- as.Date(sapply(pmquery,function(x){paste(x[1,'year'], x[1,'month'], x[1,'day'],sep="-")}))
pmquery <- pmquery[datePMID >= queryDate]
pmquery.dataframe <- ldply(pmquery,data.frame)
pmquery.dataframe$Initials <- getInitials(pmquery.dataframe$firstname)
pmquery.dataframe$ColabType <- 'Co-Author'
pmquery.dataframe <- pmquery.dataframe[!duplicated(paste(pmquery.dataframe$lastname,pmquery.dataframe$Initials)),]
pmquery.dataframe <- pmquery.dataframe[order(pmquery.dataframe$lastname,pmquery.dataframe$Initials),]