ACT IntegratedExample: Difference between revisions
Justin.jee (talk | contribs) No edit summary |
Justin.jee (talk | contribs) No edit summary |
||
| (4 intermediate revisions by the same user not shown) | |||
| Line 13: | Line 13: | ||
*Aggregation | *Aggregation | ||
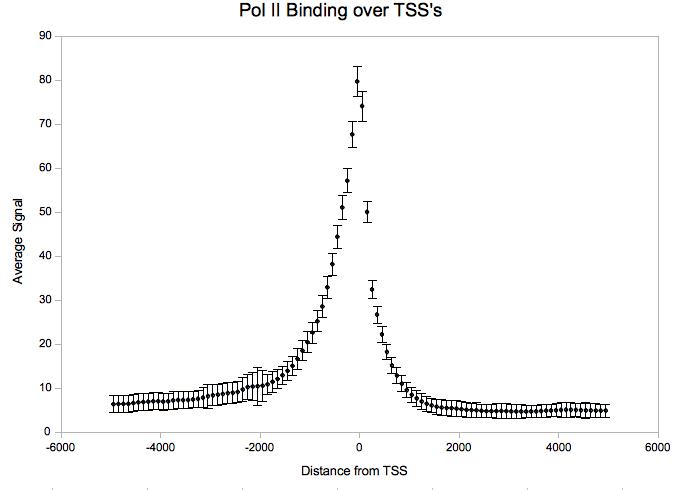
After downloading the agg-py package, it is necessary to download an annotations file with coordinates you want to aggregate over. For this example, we used all transcription start sites as taken from build 18 of the human genome annotated by the gencode project. | After downloading the agg-py package, it is necessary to download an annotations file with coordinates you want to aggregate over. For this example, we used all transcription start sites as taken from build 18 of the human genome annotated by the gencode project. For convenience/testing purposes, we have provided a parsed version of the gencode gene annotations which is compatible with act here: | ||
http://act.gersteinlab.org/gencode.coords | |||
Using ACT.py, we get a four-column file which can be plotted in Excel or using R: | Using ACT.py, we get a four-column file which can be plotted in Excel or using R: | ||
<pre>python ACT.py --nbins=50 --mbins=0 --radius=5000 gencode.coords PolII.sgr > PolII.out &</pre> | |||
[[Image:Agg_wg.png]] | [[Image:Agg_wg.png]] | ||
| Line 32: | Line 36: | ||
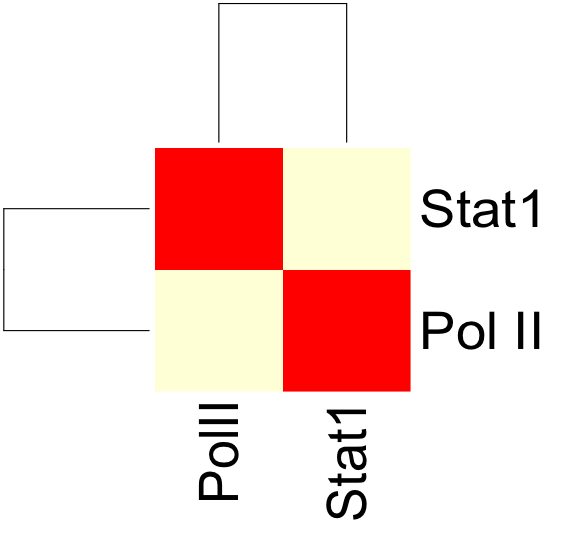
The resulting correlation matrix can be plotted in R using the heatmap function: | The resulting correlation matrix can be plotted in R using the heatmap function: | ||
<pre> | |||
#! /bin/sh | |||
lib=WEB-INF/lib | |||
cp=WEB-INF/classes/:$lib/jsci-core-1.1.jar | |||
bin=100 | |||
java -Xmx1024m -cp $cp org.gersteinlab.act.BinFileCreator config.txt Mean $bin output/PolII$bin.wig ../PolII.bedGraph4 | |||
java -Xmx1024m -cp $cp org.gersteinlab.act.BinFileCreator config.txt Mean $bin output/Stat1$bin.wig ../Stat1.bedGraph4 | |||
java -Xmx1024m -cp $cp org.gersteinlab.act.CorrelationsCalculator config.txt Pearson output/correlation_pearson.txt output/PolII$bin.wig output/Stat1$bin.wig | |||
</pre> | |||
[[Image:Corr_wg_heatmap.png]] | [[Image:Corr_wg_heatmap.png]] | ||
| Line 43: | Line 59: | ||
Go to saturation.sh and change the input files to PolII.bed and Stat1.bed. It will also probably be necessary to change the alloted memory (in the second of the final java command in saturation.sh, change the numbers in the second field from 128 to 4096). Since there are only two signal tracks, the Saturation and Correlation analyses provide overlapping information. | Go to saturation.sh and change the input files to PolII.bed and Stat1.bed. It will also probably be necessary to change the alloted memory (in the second of the final java command in saturation.sh, change the numbers in the second field from 128 to 4096). Since there are only two signal tracks, the Saturation and Correlation analyses provide overlapping information. | ||
[[Image:Saturation_wg_t20.png| | saturation.sh: | ||
<pre> | |||
#! /bin/sh | |||
lib=WEB-INF/lib | |||
cp=WEB-INF/classes/:$lib/jfreechart-1.0.13.jar:$lib/itext-1.4.3.jar:$lib/jcommon-1.0.16.jar | |||
infiles='../PolII_20.bed ../Stat1_20.bed' | |||
java -Xmx4096m -cp $cp org.gersteinlab.act.SaturationPlotsCreator output/saturation.pdf output/saturaion.txt 0 0 $infiles | |||
</pre> | |||
The pink bars correspond to the range of covered values as determined by saturating the files in every possible order. | |||
[[Image:Saturation_wg_t20.png|x500px]] | |||
Latest revision as of 18:20, 23 October 2010
ACT Integrated Example: Whole-Genome ChIP-Seq experiments
Here we describe how to use ACT on a set of two large example files from ChIP-Seq experiments.
To begin, download the signal files (from Gerstein Lab's PeakSeq project) here:
http://archive.gersteinlab.org/proj/PeakSeq/Scoring_ChIPSeq/Results/PolII/Signal_Maps/PolII/
http://archive.gersteinlab.org/proj/PeakSeq/Scoring_ChIPSeq/Results/STAT1/Signal_Maps/STAT1/
These are zipped signal files for PolII and Stat1. After unzipping the files for each transcription factor, it is useful to cat the signal tracks for the individual chromosomes together so that all the signals are in only two files. In this walkthrough they will be referred to as PolII.sgr and Stat1.sgr.
- Aggregation
After downloading the agg-py package, it is necessary to download an annotations file with coordinates you want to aggregate over. For this example, we used all transcription start sites as taken from build 18 of the human genome annotated by the gencode project. For convenience/testing purposes, we have provided a parsed version of the gencode gene annotations which is compatible with act here:
http://act.gersteinlab.org/gencode.coords
Using ACT.py, we get a four-column file which can be plotted in Excel or using R:
python ACT.py --nbins=50 --mbins=0 --radius=5000 gencode.coords PolII.sgr > PolII.out &
- Correlation
For this example, since we are dealing with signal tracks rather than SNP positions or a bed file of genomic locations, we will use the correlation tool found in corr-sat-bundle to do the correlation calculation. First, it is necessary to convert the signal tracks from sgr to wig format. A script which does this can be found here:
http://act.gersteinlab.org/sgr2wig.pl
Once we have converted both the PolII and Stat1 sgr files to wig files, we can change the parameters in correlation.sh so that the input files are "PolII.wig" and "Stat1.wig"
In addition, it is important to change config.txt so that the list of genomic regions includes all chromosomes, including mitochondrial sequences. (When the package is first downloaded, all lines except for the one denoting chromosome 22 are commented out).
Once both the correlation.sh and config.txt files have been modified, we can run correlation.sh. This will produce an output file with a correlation matrix describing the correlation coefficients between PolII and Stat1 signal.
The resulting correlation matrix can be plotted in R using the heatmap function:
#! /bin/sh lib=WEB-INF/lib cp=WEB-INF/classes/:$lib/jsci-core-1.1.jar bin=100 java -Xmx1024m -cp $cp org.gersteinlab.act.BinFileCreator config.txt Mean $bin output/PolII$bin.wig ../PolII.bedGraph4 java -Xmx1024m -cp $cp org.gersteinlab.act.BinFileCreator config.txt Mean $bin output/Stat1$bin.wig ../Stat1.bedGraph4 java -Xmx1024m -cp $cp org.gersteinlab.act.CorrelationsCalculator config.txt Pearson output/correlation_pearson.txt output/PolII$bin.wig output/Stat1$bin.wig
- Saturation
It is necessary to convert the wig files to bed files without a "signal" component. The following script takes wig files and converts them into bed files, with coordinates representing the regions of the signal track which are above a certain threshold (in this case 20).
http://act.gersteinlab.org/wig2bed.pl
Go to saturation.sh and change the input files to PolII.bed and Stat1.bed. It will also probably be necessary to change the alloted memory (in the second of the final java command in saturation.sh, change the numbers in the second field from 128 to 4096). Since there are only two signal tracks, the Saturation and Correlation analyses provide overlapping information.
saturation.sh:
#! /bin/sh lib=WEB-INF/lib cp=WEB-INF/classes/:$lib/jfreechart-1.0.13.jar:$lib/itext-1.4.3.jar:$lib/jcommon-1.0.16.jar infiles='../PolII_20.bed ../Stat1_20.bed' java -Xmx4096m -cp $cp org.gersteinlab.act.SaturationPlotsCreator output/saturation.pdf output/saturaion.txt 0 0 $infiles
The pink bars correspond to the range of covered values as determined by saturating the files in every possible order.