RSEQtools
From GersteinInfo
(→mrfQuantifier) |
(→mrfQuantifier) |
||
| Line 426: | Line 426: | ||
| - | [[Image:mrfQuantifier.png|thumb|1000px| | + | [[Image:mrfQuantifier.png|thumb|1000px|center|Determining overlaps between annotation entries and reads]] |
| - | <br> | + | <br><br> |
=== Visualization tools === | === Visualization tools === | ||
Revision as of 12:07, 1 November 2010
Contents |
Introduction
The advent of next-generation sequencing for functional genomics has given rise to quantities of sequence information that are often so large that they are difficult to handle. Moreover, sequence reads from a specific individual can contain sufficient information to potentially identify that person, raising significant privacy concerns. In order to address these issues we have developed the Mapped Read Format (MRF), a compact data summary format for both short and long read alignments that enables the anonymization of confi-dential sequence information, while allowing one to still carry out many functional genomics studies. We have developed a suite of tools that uses this format for the analysis of RNA-Seq experiments. RSEQtools consists of a set of modules that perform common tasks such as calculating gene expression values, generating signal tracks of mapped reads, and segmenting that signal into actively transcribed regions. In addition to the anonymization afforded by this format it also facilitates the decoupling of the alignment of reads from downstream analyses.
Overview
The following sections provide documenation for the modules that are part of RSEQtools (http://rseqtools.gersteinlab.org/). This documentation is intended for the end-users and can also be found at http://info.gersteinlab.org/RSEQtools. RSEQtools is implemented in C and uses a general C library called BIOS.
The full documentation for developers can be found here:
- RSEQtools: http://archive.gersteinlab.org/proj/rnaseq/doc/mrf/
- BIOS: http://archive.gersteinlab.org/proj/rnaseq/doc/bios/
Data formats
Mapped Read Format (MRF)
The Mapped Read Format (MRF) flat file consists of three components and this format is closely associated with the software components of RSEQtools
1. Comment lines. Comment lines are optional and start with a '#' character. 2. Header line. The header line is required and specifies the type of each column. 3. Mapped reads. Each read (single-end or paired-end) is represented by on line.
Required column:
* AlignmentBlocks, each alignment block must contain the following attributes: TargetName:Strand:TargetStart:TargetEnd:QueryStart:QueryEnd
Optional columns:
* Sequence * QualityScores * QueryId
Example file:
# Comments # Required field: Blocks [TargetName:Strand:TargetStart:TargetEnd:QueryStart:QueryEnd] # Optional fields: Sequence,QualityScores,QueryId AlignmentBlocks chr1:+:2001:2050:1:50 chr1:+:2001:2025:1:25,chr1:+:3001:3025:26:50 chr2:-:3001:3051:1:51|chr11:+:4001:4051:1:51 chr2:-:6021:6050:1:30,chr2:-:7031:7051:31:51|chr11:+:4001:4051:1:51 contigA:+:5001:5200:1:200,contigB:-:1200:1400:200:400
Notes:
* Paired-end reads are separated by ‘|’ * Alignment blocks are separated by ‘,’ * Features of a block are separated by ‘:’ * Columns are tab-delimited * Columns can be arranged in any order * Coordinates are one-based and closed (inclusive)
Use MRF for confidential data
It is straightforward to use MRF to separate the confidential information, i.e. the sequences, from the alignment data. The MRF file can be split in 2 files: one file can include AlignmentBlocks and QueryID, whereas a second file would can contain Sequence and QueryID. From a practical viewpoint it is also easy to create these two files.
Assuming we have the columns AlignmentBlocks, Sequence, and QueryID as column 1, 2, and 3, respectively:
$ cut -f1,3 file.mrf > alignments.mrf $ cut -f2,3 file.mrf > sequences.mrf
- alignments.mrf would contain the alignment data and the query ID; it can be freely shared since it does not include confidential information;
- sequences.mrf would contain the sequence data; which, potentially, could be used to identify an individual and thus may be subjected to more stringent rules.
Interval
The Interval format consists of eight tab-delimited columns and is used to represent genomic intervals such as genes. This format is closely associated with the intervalFind module, which is part of BIOS. This module efficiently finds intervals that overlap with a query interval. The underlying algorithm is based on containment sublists: Alekseyenko, A.V., Lee, C.J. "Nested Containment List (NCList): A new algorithm for accelerating interval query of genome alignment and interval databases" Bioinformatics 2007;23:1386-1393 [1].
1. Name of the interval 2. Chromosome 3. Strand 4. Interval start (with respect to the "+") 5. Interval end (with respect to the "+") 6. Number of sub-intervals 7. Sub-interval starts (with respect to the "+", comma-delimited) 8. Sub-interval end (with respect to the "+", comma-delimited)
Example file:
uc001aaw.1 chr1 + 357521 358460 1 357521 358460 uc001aax.1 chr1 + 410068 411702 3 410068,410854,411258 410159,411121,411702 uc001aay.1 chr1 - 552622 554252 3 552622,553203,554161 553066,553466,554252 uc001aaz.1 chr1 + 556324 557910 1 556324 557910 uc001aba.1 chr1 + 558011 558705 1 558011 558705
In this example the intervals represent a transcripts, while the sub-intervals denote exons.
Note: the coordinates in the Interval format are zero-based and the end coordinate is not included.
BED
The BED format is used to represent contiguous genomic regions. It consists of three required columns (tab-delimited).
1. Chromosome 2. Start 3. End
Example file:
chr1 1000 5000 chr3 500 600 chrX 4000 4250
Full documentation can be found at UCSC
Note: the coordinates in the BED format are zero-based and the end coordinate is not included.
WIG
The WIG format is used to represent dense and continuous genomic data. There are two options for formatting wiggle data: variableStep and fixedStep.
In the context of RSEQtools, the variable step formatting is used and only positions with non-zero values are represented.
Example file:
track type=wiggle_0 name="test_chr22" variableStep chrom=chr22 span=1 17535712 1.67 17535713 1.67 17535714 1.67 17535715 1.67 17535716 1.67
Full documentation can be found at UCSC
Note: the coordinates in the WIG format are zero-based.
GFF
The GFF format is used to describe genes and other features. It consists of nine tab-delimited columns.
1. Name 2. Source 3. Feature 4. Start 5. End 6. Score 7. Strand 8. Frame 9. Group
Example file:
browser hide all track name="chr11" visibility=2 chr11 MRF feature 46772115 46772161 . - . TG5 chr11 MRF feature 46772668 46772695 . - . TG5 chr11 MRF feature 118521207 118521252 . + . TG21 chr11 MRF feature 118526315 118526343 . + . TG21
Full documentation can be found at UCSC
Note: the coordinates in the BED format are one-based and the end coordinate is included.
PSL
The PSL format represents alignments from the BLAT alignment program.
Full documentation can be found at UCSC
SAM
SAM (Sequence Alignment/Map) format is a generic format for storing large nucleotide sequence alignments.
Full documentation can be found at SAMtools
List of programs
This is the documentation for the end-users. The full documentation for developers can be found here.
Format conversion utilities
The following programs convert the output from various alignment programs into MRF.
bowtie2mrf
bowtie2mrf converts read alignments from Bowtie into MRF.
Usage:
bowtie2mrf <genomic|junctions|paired> [-sequence] [-qualityScores] [-IDs]
- Inputs: Takes Bowtie output from STDIN
- Outputs: Reports MRF to STDOUT
- Required arguments
- genomic - convert single-end reads that were aligned against a genomic reference sequence using Bowtie
- junctions - convert single-end reads that were aligned against a splice junction library (generated by createSpliceJunctionLibrary) using Bowtie
- paired - convert paired-end reads that were aligned using Bowtie
- Optional arguments
Note: bowtie2mrf assumes that bowtie was run using the default option for the -B parameter
-B/--offbase <int> leftmost ref offset = <int> in bowtie output (default: 0)
psl2mrf
psl2mrf converts read alignments from BLAT into MRF.
Usage:
psl2mrf
- Inputs: Takes BLAT alignments in PSL format from STDIN
- Outputs: Reports MRF to STDOUT
- Required arguments
- None
- Optional arguments
- None
singleExport2mrf
singleExport2mrf converts single-end read alignments from ELAND (export file) into MRF.
Usage:
singleExport2mrf
- Inputs: Takes ELAND single-end alignments in export format from STDIN
- Outputs: Reports MRF to STDOUT
- Required arguments
- None
- Optional arguments
- None
sam2mrf
sam2mrf converts SAM format into MRF
Usage:
sam2mrf
- Inputs: Takes SAM format from STDIN
- Outputs: Reports MRF to STDOUT
- Required arguments
- None
- Optional arguments
- None
Please note that for paired-end data, sam2mrf requires the mate pairs to be on subsequent lines. You may want to sort the SAM file first.
Example:sort -r file.sam | sam2mrf > file.mrf
Genome annotation tools
The following tools are helpful in manipulating annotation files.
createSpliceJunctionLibrary
This program is used to create a splice junction library from an annotation set. It creates all pair-wise splice junctions within a transcript.
Usage:
createSpliceJunctionLibrary <file.2bit> <file.annotation> <sizeExonOverlap>
- Inputs: None
- Outputs: Reports the slice junctions in FASTA format
- Required arguments
- file.2bit - genome reference sequence in 2bit format
- file.annotation - annotation set in Interval format (each line represents one transcript)
- sizeExonOverlap - defines the number of nucleotides included from each exon
- Optional arguments
- None
Example output:
>chr1|12162|12612|65 AGGCATAGGGGAAAGATTGGAGGAAAGATGAGTGAGAGCATCAACTTCTCTCACAACCTAGGCCAGTGTGTGGTGATGCCAGGCATGCCCTTCCCCAGCATCAGGTCTCCAGAGCTGCAGAAGACGACGG >chr1|12162|13220|65 AGGCATAGGGGAAAGATTGGAGGAAAGATGAGTGAGAGCATCAACTTCTCTCACAACCTAGGCCAGCAGGGCCATCAGGCACCAAAGGGATTCTGCCAGCATAGTGCTCCTGGACCAGTGATACACCCGG >chr1|12656|13220|65 CAGAGCTGCAGAAGACGACGGCCGACTTGGATCACACTCTTGTGAGTGTCCCCAGTGTTGCAGAGGCAGGGCCATCAGGCACCAAAGGGATTCTGCCAGCATAGTGCTCCTGGACCAGTGATACACCCGG
The identifier for each splice junction consists of four items:
1. Chromosome 2. Start position (with respect to the "+", zero-based) of the splice junction within the first exon 3. Start position (with respect to the "+", zero-based) of the splice junction within the second exon 4. Size of the exon overlap
Note: Internally the program uses twoBitToFa (part of BLAT package). Thus, the executable must be in the PATH.
mergeTranscripts
Module to merge a set of transcripts from the same gene.
Obtain unique exons from various transcript isoforms based on:
- longest isoform
- composite model (union of the exons from the different transcript isoforms)
- intersection (intersection of the exons of the different transcript isoforms)
Usage:
mergeTranscripts <knownIsoforms.txt> <file.annotation> <longestIsoform|compositeModel|intersection>
- Inputs: None
- Outputs: Reports a new annotation set of merged transcripts in Interval format
- Required arguments
- knownIsoforms.txt - file that determines which transcript isoforms belong together (see format below)
- file.annotation - annotation set in Interval format (each line represents one transcript)
- < longestIsoform | compositeModel | intersection > - determines how transcript isoforms are selected/merged:
- Optional arguments
- None
The file knownIsoforms.txt should have two columns (tab-delimited) and no header:
1. ID (int). Transcripts with the same id belong to the same gene. 2. Name of the transcript.
Example:
1 uc009vip.1 1 uc001aaa.2 2 uc009vis.1 2 uc001aae.2 2 uc009viu.1 2 uc009vit.1
interval2sequences
Module to retrieve genomic/exonic sequences for an annotation set.
Usage:
interval2sequences <file.2bit> <file.annotation> <exonic|genomic>
- Inputs: None
- Outputs: Reports the extracted sequences in FASTA format
- Required arguments
- file.2bit - genome reference sequence in 2bit format
- file.annotation - annotation set in Interval format (each line represents one transcript)
- < exonic | genomic > - exonic means that only the exonic regions are extracted, while genomic indicates that the intronic sequences are extracted as well
- Optional arguments
- None
Gene expression analysis
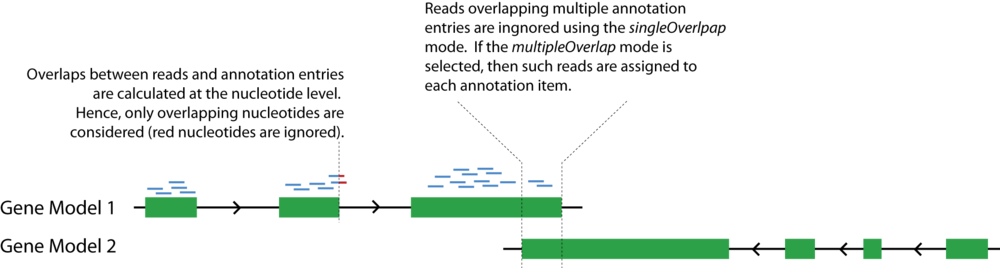
mrfQuantifier
Module to calculate gene expression values (RPKM).
Usage:
mrfQuantifier <file.annotation> <singleOverlap|multipleOverlap>
- Inputs: Takes MRF from STDIN.
- Outputs: Reports the gene expression values to STDOUT in a two-column format (tab-delimited). The first column refers to the name of the annotated feature. The second column refers to the gene expression values (RPKM; read coverage normalized per million mapped nucleotides and the length of the gene model per kb [see note below]). The output is sorted by the first column.
- Required arguments
- file.annotation - annotation set in Interval format (gene expression measurements: one line per gene model; exon expression measurements: one line per exon)
- < singleOverlap | multipleOverlap > - singleOverlap: reads that overlap with multiple annotated features are ignored; multipleOverlap: reads that overlap with multiple annotated features are counted multiple times.
- Optional arguments
- None
Note: All counts are performed at the nucleotide level. For example, if a read partially overlaps with an exon of a gene model, then only the overlapping nucleotides are counted. Therefore, the normalization is also done at the nucleotide level.
Visualization tools
The following programs are useful for converting MRF into data formats that can be viewed in a genome browser.
mrf2wig
Generates signal track (WIG) of mapped reads from a MRF file. By default, the values in the WIG file are normalized by the total number of mapped reads per million. Only positions with non-zero values are reported.
Usage:
mrf2wig <prefix> [counts]
- Inputs: Takes MRF from STDIN.
- Outputs: Generates a WIG file for each chromosome occurring in the MRF input file.
- Required arguments
- prefix - specifies the prefix used to generate the output files. The following naming convention is used: prefix_chrXXX.wig
- Optional arguments
- counts - do not normalize values, report raw counts int the WIG file instead
mrf2gff
Generates a GFF file of mapped splice junction reads from a MRF file.
Usage
mrf2wig <prefix>
- Inputs: Takes MRF from STDIN.
- Outputs: Generates a GFF file for each chromosome occurring in the MRF input file.
- Required arguments
- prefix - specifies the prefix used to generate the output files. The following naming convention is used: prefix_chrXXX.gff
- Optional arguments
- None
mrf2bedGraph
Module to convert MRF to BedGraph. Generates a BedGraph, where the counts are normalized by the total number of mapped reads per million.
Usage:
mrf2bedGraph <prefix> [<trackName>]
- Inputs: Takes MRF from STDIN.
- Outputs: Generates a BedGraph file for each chromosome occurring in the MRF input file.
- Required arguments
- prefix - specifies the prefix used to generate the output files. The following naming convention is used: prefix_chrXXX.bgr
- Optional arguments
- trackName - by default, the prefix is used as track name. If trackName is specified, then this string will be used instead.
Segmentation of mapped reads
wigSegmenter
Module to segment a WIG signal track using the maxGap-minRun algorithm. The output is a set of transcriptionally active regions (TARs) in BED format.
Usage:
wigSegmenter <wigPrefix> <threshold> <maxGap> <minRun>
- Inputs: None
- Outputs: Generates a BED file for each chromosome occurring in the MRF input file.
- Required arguments
- Optional arguments
- None
Annotation statistics tools
The following modules are useful for calculating annotation statistics given a set of mapped reads.
mrfAnnotationCoverage
Module to calculate annotation coverage. Sample a set of mapped reads and determine the fraction of transcripts (specified in file.annotation) that have at least <coverageFactor>-times uniform coverage.
Usage:
mrfAnnotationCoverage <file.annotation> <numTotalReads> <numReadsToSample> <coverageFactor>
- Inputs: Takes MRF from STDIN
- Outputs: Reports the fraction of transcripts that have at least <coverageFactor>-times uniform coverage to STDOUT
- Required arguments
- Optional arguments
- None
mrfMappingBias
Module to calculate mapping bias for a given annotation set. Aggregates mapped reads that overlap with transcripts (specified in file.annotation) and outputs the counts over a standardized transcript (divided into 100 equally sized bins) where 0 represents the 5' end of the transcript and 1 denotes the 3' end of the transcripts. This analysis is done in a strand specific way.
Usage:
mrfMappingBias <file.annotation>
- Inputs: Takes MRF from STDIN
- Outputs: Outputs the number of mapped reads for each bin of the standardized transcript to STDOUT
- Required arguments
- file.annotation - annotation set in Interval format (each line represents one transcript)
- Optional arguments
- None
MRF selection utilities
The following utilities are helpful to select subsets of an MRF file.
mrfSampler
Randomly select a subset of MRF entries.
Usage:
mrfSampler <proportionOfReadsToSample>
- Inputs: Takes MRF from STDIN
- Outputs: Outputs MRF to STDOUT
- Required arguments
- proportionOfReadsToSample - fraction of reads to sample (on average)
- Optional arguments
- None
mrfSelectRegion
Select reads that overlap with a specified genomic region.
Usage:
mrfSelectRegion <targetName:targetStart-targetEnd>
- Inputs: Takes MRF from STDIN
- Outputs: Outputs MRF to STDOUT
- Required arguments
- targetName:targetStart-targetEnd - region of interest
- Optional arguments
- None
mrfSelectSpliced
Select reads that span a splice junction.
Usage:
mrfSelectSpliced
- Inputs: Takes MRF from STDIN
- Outputs: Outputs MRF to STDOUT
- Required arguments
- None
- Optional arguments
- None
Note: The purpose of mrfSelectSpliced is to extract mapped reads that align to a splice junction from an existing MRF file. It is important to note that this utility is not used to convert the output of a specific mapping program.
mrfSubsetByTargetName
Split up an MRF file by chromosome.
Usage:
mrfSubsetByTargetName <file.mrf[.gz]>
- Inputs: None
- Outputs: Outputs a separate MRF file for each chromosome.
- Required arguments
- file.mrf[.gz] - The MRF file to split up. The file can be gzipped (optional).
- Optional arguments
- None
mrfSelectAnnotated
Module to select a subset of reads that overlap with a specified annotation set.
Usage:
mrfSelectAnnotated <file.annotation> <include|exclude>
- Inputs: Takes MRF from STDIN
- Outputs: Outputs MRF to STDOUT
- Required arguments
- file.annotation - annotation set in Interval format (one transcript per line).
- < include | exclude > - include: report reads that overlap with exonic regions of the annotation set; exclude: report reads that do not overlap with exonic regions of the annotation set
- Optional arguments
- None
Auxiliary utilities
This section includes various data format conversion utilities.
bed2interval
Utility to convert BED format into Interval format.
Usage:
bed2interval
- Inputs: Takes data in BED format from STDIN
- Outputs: Outputs data in Interval format to STDOUT
- Required arguments
- None
- Optional arguments
- None
interval2bed
Utility to convert Interval format into BED format.
Usage:
interval2bed
- Inputs: Takes data in Interval format from STDIN
- Outputs: Outputs data in BED format to STDOUT
- Required arguments
- None
- Optional arguments
- None
interval2gff
Utility to convert Interval format into GFF format.
Usage:
interval2gff <trackName>
- Inputs: Takes data in Interval format from STDIN
- Outputs: Outputs data in GFF format to STDOUT
- Required arguments
- trackName - track name used in the GFF file
- Optional arguments
- None
gff2interval
Utility to convert GFF format into Interval format.
Usage:
gff2interval
- Inputs: Takes data in GFF format from STDIN
- Outputs: Outputs data in Interval format to STDOUT
- Required arguments
- None
- Optional arguments
- None
export2fastq
Module to generate FASTQ sequences from an ELAND export file.
Usage:
export2fastq
- Inputs: Takes an ELAND export file from STDIN
- Outputs: Reports the extracted sequences in FASTQ format to STDOUT
- Required arguments
- None
- Optional arguments
- None
mrf2sam
Usage:
mrf2sam
- Inputs: Takes MRF from STDIN
- Outputs: Outputs SAM to STDOUT
- Required arguments
- None
- Optional arguments
- None