CRIT
From GersteinInfo
(→Cross Pattern Identification Technique (CRIT)) |
(→Cross Pattern Identification Technique (CRIT)) |
||
| (One intermediate revision not shown) | |||
| Line 1: | Line 1: | ||
=Cross Pattern Identification Technique (CRIT)= | =Cross Pattern Identification Technique (CRIT)= | ||
| + | |||
| + | ==Algorithm Overview== | ||
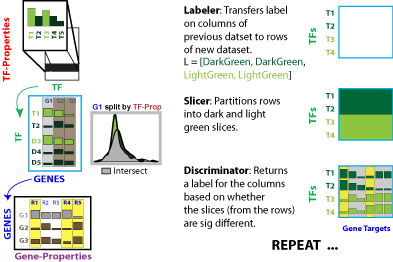
Label, Slice, Discriminate, Repeat. | Label, Slice, Discriminate, Repeat. | ||
| - | [[File:alg.png | + | [[File:alg.png]] |
Latest revision as of 16:38, 29 January 2011
Contents |
Cross Pattern Identification Technique (CRIT)
Algorithm Overview
Label, Slice, Discriminate, Repeat.
Core Functions and Their Parameters
Initializer
Initializer: Only run the first time to obtain some set of labels for the rows of A. In all remaining steps, the discriminator generates the new label for propogation. Alternatively, this step can be skipped and a label can be supplied directly to the labeler function.
Required Arguments: Column of A, type of partitioning
Output: Vector assigning a label to every ROW of Column of A
initializer<-function(A, type=c("median","mean")) {
#Create empty vector with the same number of rows of A
label<-matrix(0, length(A))
#Get value to threshold off of
t<-getThreshold(A, type=type)
#Create label
label<-labelSlicer(A ,t)
return (label)
}
Labeler
In implementation both labeler and slicer (labelSlicer) are integrated as the label is a simple column vector. We show the breakdown to make the connection between the algorithm design and implementation more transparent.
Labeler: Transfers labels on columns of previous datasets (A) to rows of new dataset (B)
Required Arguments: Matrix (B), vector assigning a label to every ROW of A
Output: Vector assigning a label to every COLUMN of B
Slicer
Slicer: Partition rows of N into "slices" based on labels from A
Required Arguments: Matrix (B), Vector assigning a label to every COLUMN of B
Output: Set of slices where slice is defined as a set of rows of B all containing the same label (in practice this is just the grouping variable as opposed to actual data slices)
labelSlicer<-function(values, t){
#Set those strictly greater than threshold to 1
index<-which(values>=t)
label(index)<-1
return (label)
}
Discriminator
Discriminator: Evaluates the discriminatory power of labels generated from B in the context of C. Set of slices where slice is defined as a set of rows of B all containing the same label. A random label would show no difference in the slices. (In practice this is implemented as an index instead of the literal data slice).
Required Arguments: Matrix (B), label
Optional Arguments: set to TRUE to compute the FDR
Output: Value of test for every column of B
#In practice this is implemented as a index
#Additional discriminator functions that use KS test, hypergeometric
#distribution, etc are possible
discriminator<-function(mat=B, label=l, multCorrect=FALSE){
#Create empty list
ttest_struct<-list();
#Compute value of test for each row
for(i in 1:nrow(X)){
temp<-t.test(as.numeric(B[i,])~label)
ttest_struct[[i]]=temp;
}
#Extract pvalue from structure
all_pval<-getPvalfromStruct(ttest_struct);
#Test if we want to compute FDR
if (multCorrect) { all_pval<-p.adjust(all_pval, method="fdr") }
return (all_pval)
}
AUXILLIARY FUNCTIONS
Checking against Threshold
Returns a value to split on for the base case
getThreshold<-function(A, type=c("median","mean")) {
type <- match.arg(type)
threshold <- switch(type,
mean = mean(A),
median = median(A)
)
return (threshold)
}
Return t-test Object
Returns only the t-test from the ttest object
getTfromStruct<-function(struct){
p<-unlist(lapply(struct, "[",1));
return(p)
}
Return p Value Object
Returns only the pvalues from the ttest object
getPvalfromStruct<-function(struct){
p<-unlist(lapply(struct, "[",3));
return(p)
}