What is a gene?
From GersteinInfo
(Created page with '''``The gene is a union of genomic sequences encoding a coherent set of potentially overlapping functional products."'' ''from'': <br> '''What is a gene, post-ENCODE? History an…')
Latest revision as of 13:05, 10 June 2010
``The gene is a union of genomic sequences encoding a coherent set of potentially overlapping functional products."
from:
What is a gene, post-ENCODE? History and updated definition
MB Gerstein, C Bruce, JS Rozowsky, D Zheng, J Du, JO Korbel, O Emanuelsson, ZD Zhang, S Weissman M Snyder (2007) Genome Research, 17, 669-681.
Free Full Text | PMID: 17567988
Contents |
Why a new definition for the term ``gene" is needed?
The classical view of a gene as a discrete element in the genome has been shaken by ENCODE
The ENCODE consortium recently completed its characterization of 1% of the human genome by various high-throughput experimental and computational techniques designed to characterize functional elements. This project represents a major milestone in the characterization of the human genome, and the current findings show a striking picture of complex molecular activity.
Before the advent of the ENCODE project, there were a number of aspects of genes that were very complicated, but much of this complexity was in some sense swept under the rug and did not really affect the fundamental definition of a gene. The experience of the ENCODE project, particularly the mapping of transcriptional activity and regulation using tiling arrays, has extended these puzzling and confusing aspects of genes, bringing them to the forefront, where one has to grapple more directly with them in relation to the definition of what a gene is.
What the ENCODE experiments show: Lattices of long transcripts and dispersed regulation
- Unannotated transcription
- Unannotated and alternative TSSs
- More alternative splicing
- Dispersed regulation
- Noncoding RNAs
- Pseudogenes
At this point, it is not clear what to do: In the extreme, we could declare the concept of the gene dead and try to come up with something completely new that fits all the data. However, it would be hard to do this with consistency. Here, we made a tentative attempt at a compromise, devising updates and patches for the existing definition of a gene.
How does the new definition reflect a shift in scientific thinking?
The classical view of a gene as a unit of hereditary information aligned along a chromosome, each coding for one protein, has changed dramatically over the past century. For Morgan, genes on chromosomes were like beads on a string. The molecular biology revolution changed this idea considerably. And now the ENCODE project has increased the complexity still further.
An important aspect of our proposed definition is the requirement that the protein or RNA products must be functional for the purpose of assigning them to a particular gene. We believe this connects to the basic principle of genetics, that genotype determines phenotype. At the molecular level, we assume that phenotype relates to biochemical function. Our intention is to make our definition backwardly compatible with earlier concepts of the gene.
What does leaving regulatory sequences out of the definition mean?
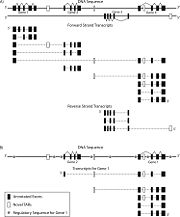
Although regulatory regions are important for gene expression, we suggest that they should not be considered in deciding whether multiple products belong to the same gene. This aspect of the definition results from our concept of the bacterial operon. The fact that genes in an operon share an operator and promoter region has traditionally not been considered to imply that their protein products are alternative products of a single gene. Consequently, in higher eukaryotes, two transcripts that originate from the same transcription start site (sharing the same promoter and regulatory elements) but do not share any sequence elements in their final products (e.g., because of alternative splicing) would not be products of the same gene. A similar logic would apply to multiple transcripts sharing a common but distant enhancer or insulator. Regulation is simply too complex to be folded into the definition of a gene, and there is obviously a many-to-many (rather than one-to-one) relationship between regulatory regions and genes.
How is this new definition for genes relevant to the general public?
This emphasis of our new definition on functional products, of course, highlights the issue of what biological function actually is. With this, we move the hard question from "what is a gene?" to "what is a function?" We expect this functional view of genes to achieve better public perception, since it focuses on the biological processes the genes are involved in, which when described at a certain level would be much easier for the public to understand compared to the structure of those genes.
High-throughput biochemical and mutational assays will be needed to define function on a large scale. Hopefully, in most cases it will just be a matter of time until we acquire the experimental evidence that will establish what most RNAs or proteins do. Until then we will have to use "place-holder" terms like TAR, or indicate our degree of confidence in assuming function for a genomic product. We may also be able to infer functionality from the statistical properties of the sequence.